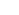Genetic structure in natural populations of Dacrydium elatum in the Central Highlands of Vietnam inferred by microsatellite molecular marker (SSR)
20/07/2021Assessment of genetic diversity and structure among populations and within species is important for genetic conservation and plant breeding programs. Genetic diversity is also important for a species' adaptability for long-term survival and adaptation to changing environmental conditions. Therefore, a better understanding of the genetic process and genetic diversity of rare and endangered species will provide important data for the development of conservation and sustainable use strategies.
Dacrydium elatum (Roxb.) Wall.) is one of 33 species of conifers native to Vietnam, growing in tropical humid evergreen closed forests at altitudes from 700m to 2,000m. In the wild, this species is distributed in provinces of Ha Giang, Tuyen Quang, Quang Ninh, Lai Chau, Ha Tinh, Quang Binh, Thua Thien Hue, Da Nang, Kon Tum, Gia Lai, Dak Lak, Lam Dong, and Kien Giang and on the list of endangered species of Vietnam Red Book (2007). So far, many populations of Dacrydium elatum are threatened due to overexploitation, deforestation, or managers and scientists’ lack of important information on genetic diversity and structure of natural populations of this species.

Natural pure-species Dacrydium elatum population in Kon Chu Rang Nature Reserve, Gia Lai and Collection and processing of specimens in the field
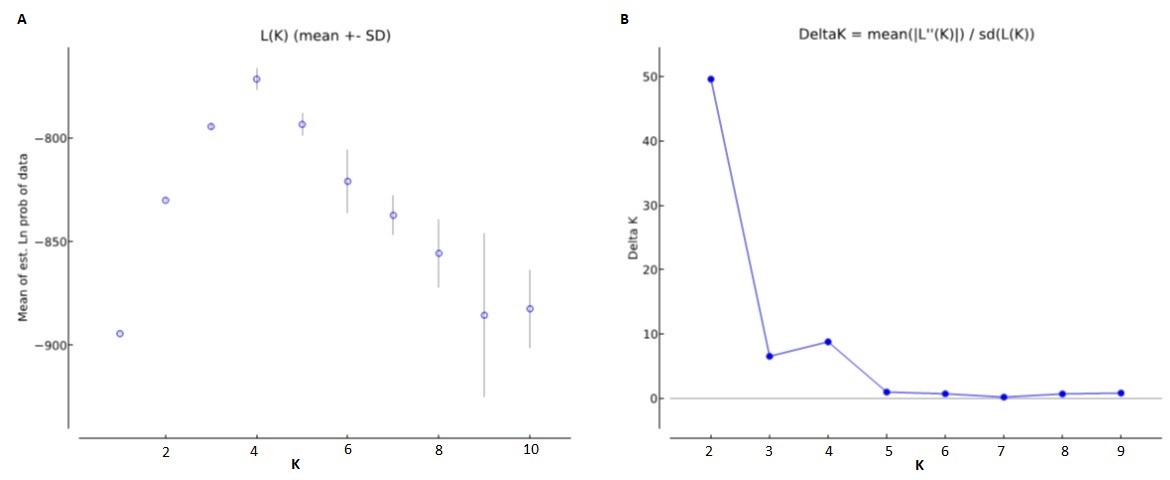
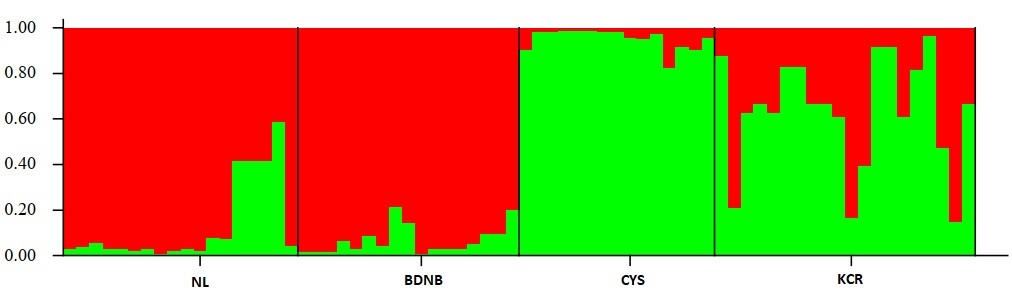
In this study, 8 SSR markers were used to assess the genetic diversity and structure of 80 natural individuals from 4 populations in Ngoc Linh (NL) Nature Reserve, Kon Chu Rang (KCR) Nature Reserve, Chu Yang Sin (CYS) National Park, and Bidoup-Nui Ba (BDNB) National Park. The results showed that the genetic diversity among the population was quite high with the observed heterozygous gene coefficient (Ho) and the expected heterozygous gene coefficient (He) of 0.555 and 0.429, respectively. The Shannon genetic diversity index (I) was 0.618 and genetic variation among populations was low (Fst = 0.097). Among which, the BDNB population has the highest level of genetic diversity compared to the other three populations (NL, CYS, and KCR). Analysis of molecular variance (AMOVA) revealed high genetic variation among individuals within a population (87%) compared to that among populations (13%). The UPGMA-based Tree Diagram showed the ramifications were largely consistent with pedigree and geographical origin. The three populations of NL, KCR, and BDNB were grouped together and the CYS population was segregated. In population structure analysis, the largest ΔK value was determined when K=2, showing that 80 individuals in 4 populations are divided into two main gene groups. The first gene group was dominant in two populations (NL and BDNB) and the second gene group consisted of two populations (CYS and KCR).Conservation and management of a species requires information on the ecology and genetic diversity. Currently, to develop appropriate conservation strategies, experts often use microsatellite molecular markers (simple sequence repeats - SSR) to assess genetic diversity and structure both at the population and species level. SSR is characterized by its polyallelic nature, co-dominant inheritance, repeatability, and widespread presence in the whole genome, making it an ideal indicator for the exploration of population genetic diversity and structure, and wide range of applications. In Vietnam, although previous studies have assessed genetic variation and confirmed taxonomic status at the molecular level of species of D. pectinatum, D. imbricatus, D. elatum, the genetic structure of Dacrydium elatum has not been fully studied. Meanwhile, to develop strategies for conservation and sustainable use of species, understanding genetic diversity is crucial. Therefore, the objective of this study is to determine the diversity and genetic struture of Dacrydium elatum populations in the Central Highlands and provide guidelines for conservation, management, and restoration.
Population genetic structure of the endangered Dacrydium elatum in Vietnam
Overall, presently at the population level, Dacrydium elatum maintains a moderate level of genetic diversity and, among populations, has low levels of genetic variation, but high intra-individual genetic one. The habitats of this conifer have been greatly affected by human activity and commercial logging in recent decades since 1980. All studied populations of this species are located in isolated small forests, unsuitable for their survival. Based on our results, conservators should also undertake ex-situ conservation activities to avoid future inbreeding. This study contributes to providing a theoretical basis for genetic resources management and identification of Dacrydium elatum and a scientific basis for breeding.
Sources: Dinh Duy Vu, Quoc Khanh Nguyen and Mai Phuong Pham, 2021. Genetic structure in natural populations of Dacrydium elatum (Roxb.) Wall. (Podocarpaceae) in the Central Highlands of Vietnam inferred by Microsatellites. E3S Web of Conferences 265, 01030 (2021). https://doi.org/10.1051/e3sconf/202126501030
Article and photo by Dr. Vu Dinh Duy/Institute of Tropical Ecology